Introduction
As the need to better our understanding of the biodiversity of the world’s various species increases, it becomes a prerequisite to conducting essential testing to have a clear understanding of the various traits of the different species. Effective lab assays done through morphological and biochemical tests and modern molecular techniques have been essential for accurately identifying species and their genetic sequences and precisely identifying the many bacterial genera present in a sample. Genotypic, proteotypic, and phenotypic methods are used in bacterial identification to identify their rRNA, determine their ribosomal proteins constituents, and acknowledge their observable characteristics, such as the biochemical attributes of the bacteria, respectively. Among the most compelling contemporary bacterial identification methods involve techniques such as the molecular biology approach, mass spectrometry, and biochemical tests1. Efficient bacteriological laboratory techniques have been used in the project to identify the unknown bacteria by gram testing to identify the unknown bacteria as gram-positive or gram-negative.
This project aims to identify the unknown bacteria #55 by analyzing the chemical and physical properties of the specimen provided. This gram testing is the preliminary step of bacterial identification and subsequent classification. In order to accurately identify the organism, a gram test was the prerequisite step. The gram test analyzed the organism’s cell walls to determine if they were positive or negative. Furthermore, catalase, sucrose, lactose, starch, and urease tests were done in the second week. Lastly, a final PR mannitol test was done in the third week to test the fermentation ability of the specimen in sugar. After these tests, the unknown bacteria was identified as staphylococcus aureus.
Methods
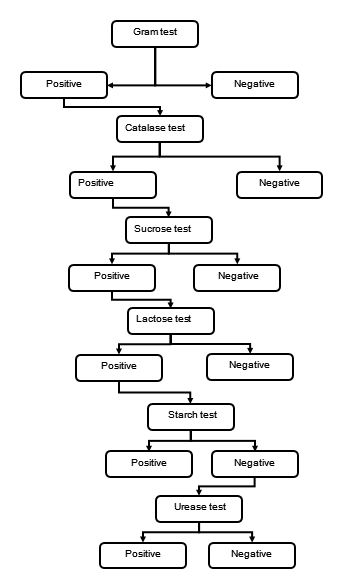
Methods Description
Gram Staining
Samples were smeared with a loopful on clean, grease-free slides. Then we air-dried them and treated the sample to a bit of heat fixing. Crystal Violet reagent was further added. The sample was kept for one minute and then rinsed with distilled water. We further flooded the gram’s iodine and waited 1 minute before rinsing with clean distilled water. We further washed the slide with the sample with acetone for 30 seconds. The sample was then kept in Safari solution for one minute before washing it with clean distilled water. Further, we air-dried the sample properly. The sample was then observed under a microscope.
Catalase Test
We used the tube method to test the unknown bacteria in the catalase test. We added 2 ml of hydrogen peroxide to a solution in a test tube. We then used a glass rod and sampled the unknown organism sample. Then the glass rod bearing the sample of the microorganism was dipped into the hydrogen peroxide solution. We observed immediately for any effervescence.
Sucrose Test
In this process, no reagents were added. An inoculated sample of the unknown organism was added aseptically to a sterilized glass vial of phenol red sucrose solution. The inoculation tube was then incubated for an additional 24 hours at a temperature of 35 to 37 C, followed by an assessment of the outcomes. The color change was observed.
Lactose Test
No reagents were added during this test. Aseptically, an inoculum was transferred from the sample of the unknown organism to a sterile tube containing phenol red lactose solution. An additional control tube was inoculated. The inoculation tube was then incubated for an additional four days at 35o C–37o C. We kept track of the color changes on the tube inoculated with the sample in contrast to the control tube daily.
Starch Test
A sample of the unknown organism was put on a sterilized labeled plate, and the inoculated plates were further incubated for two days at a stabilized 37o C. After incubation, we added iodine solution to the sample on the plates for 30 seconds and poured off the excess iodine solution. Observations were made and recorded.
Urease Test
After preparing the area to create a culture media, a sample of the unknown organism was added by a streak on the surface of the urea agar. The tube was incubated for two days at 35°–37°C in a well-aerated room with the cap left unfastened. Daily observations of the color change were recorded.
RESULTS
Gram Test
the specimen was tested for gram straining using Crystal Violet, iodine, and safari solutions and observed under a microscope. The color change was to purple.
Catalase Test
The unknown organism was tested by using a hydrogen peroxide solution. Effervescence was observed.
Sucrose Test
The unknown organism was tested by using a phenol red sucrose solution. The color of the inoculated sample changed to orange.
Lactose Test
The unknown organism was tested by using a phenol red sucrose solution. The color of the inoculated tube changed to orange against that of the control tube.
Starch Test
The unknown organism was tested by using an iodine solution. The color of the sample remained black.
Urease Test
The unknown organism was tested by using a urea agar culture. The bright pink color was observed.
Discussion
The gram-positive bacterium Staphylococcus aureus brings on several clinical illnesses. They have a cocci form and are typically grouped. These bacteria cause several types of infections in people, including bacteremia, infective endocarditis, and skin and soft tissue infections, and are among the most prevalent bacterial diseases in humans. The kind of infection and the existence or absence of drug-resistant strains significantly affect how S. aureus infections are treated. When antimicrobial therapy is required, the kind of infection and other criteria have a significant role in determining the course and form of treatment. Since preventing S. aureus infections is still challenging, attempts have centered on infection control measures such as hospital decontamination and public education on appropriate handwashing.
Conclusion
This project has been of great deliberation in identifying the evolutionary staphylococcus aureus bacterium. Analyzing staphylococcus aureus’s chemical and physical properties becomes an essential starting point for this bacterium’s classification and brings forth a better understanding of the bacteria. Understanding the organism forms the basis of analyzing its physical and biochemical composition, which is essential for today’s scientists’ efforts to understand this bacterium. In the future, there is a need to delve deeper into research beyond the contemporary understanding of this species to develop effective treatment and control measures against this organism.
References
- Talamantes-Becerra B, Georges A, Gahan ME, Kennedy K, Carling J. Identify bacterial isolates from a public hospital in Australia using complexity-reduced genotyping. Journal of microbiological methods. https://pubmed.ncbi.nlm.nih.gov/30894330/. Published 2019. Accessed December 1, 2022.
- Taylor TA, Unakal CG. Staphylococcus aureus. National Center for Biotechnology Information. https://pubmed.ncbi.nlm.nih.gov/28722898/. Published 2021. Accessed December 1, 2022.
- Knapp S. Staphylococcus aureus – the definitive guide. Biology Dictionary. https://biologydictionary.net/staphylococcus-aureus/. Published May 17, 2020. Accessed December 1, 2022.
 write
write