Abstract
The examination of environmental DNA (eDNA) has been applied to detect numerous species within an aquatic environment. Using eDNA, this experiment examines the presence or absence of the tremendous crested Newt in two distinct ponds at three year periods. From April to June, it was feasible to identify great crested newt DNA in some monthly water specimens taken from two sample ponds and ditches based on the data presented in the results. Similar levels of great crested newt eDNA (i.e., highly identification) were detected throughout March, indicating that the current survey timeframe may be extended. To determine the applicability of these findings to ponds throughout the remainder of the United Kingdom, additional research should be conducted in multiple ponds over multiple seasons. In two ponds, the current study demonstrates the effectiveness and reliability of eDNA detection in finding evidence of great crested newts.
Introduction
Animals and human beings have unique DNA, and they shed them each time in different ways, including hairs, secretions, fingerprints, shedding, and all the body parts left out in the environment. These can be collected from the environment and, by laboratory means and processes, including PCR, can be used to identify their presence in the place at a particular time. Environmental DNA (eDNA) is present in many forms, such as feces, urine, pieces of skin or hair, living or dead and degraded cells, mucus, eggs, and sperm (Laramie, 2013; Wilson & Wright, 2014) and can be sampled from aquatic environments, soil, sediment, or permafrost (Pilliod et al., 2013; Jerde & Mahon, 2015; Thomsen & Willerslev, 2015).
The study aimed to detect the presence of native species, the great crested Newt, protected in Great Britain as an endangered species in Wicken fen. The presence of the invasive fungal pathogen of amphibians was also studied.
Newts are amphibians, breeding in ponds during the spring and spending most of the year feeding on invertebrates in woodland, hedgerows, marshes, and tussocky grassland. (The wildlife trust, online) pointed at finding out if the endangered species was present in the Wicken Fen at a particular point in time, bearing in mind that they have significantly declined in population and there is a strict law for their conservation.
We used readily provided samples collected from two ponds, two ditches, and a private pond confirmed to have the species: all from the Wicken Fen Natural Reserve. A DNA extraction process was conducted, and in the first session, the primary sample was homogenized, the sub-sample concentrated then the specimen was digested. In the second part, DNA was bound to the column by attaching it to a prepared white silica gel membrane. The DNA was washed to rinse while attached to the column and then separated from the column by resuspending in the buffer. Reaction mixes for each of the two markers, sufficient for all the extracts, and the negative and positive control were prepared. The samples were then run in thermocyclers, and results were obtained after PCR.
Method
The practical was conducted procedurally, starting with sampling, DNA extraction, PCR to amplify the markers, then the electrophoresis process and visualization of the image results obtained.
Samples were collected from two ponds, two ditches, and a private pond positive for the species. The collection was done on three different dates of the year. Samples were collected from different pond parts, homogenized, and divided into different samples for different groups. Each group had its samples labeled appropriately for identification.
Each group had nine samples and a negative control of double distilled water, making it ten. Centrifuging, separations, and repetition were done to concentrate the samples. ATL buffer was included in the process.
Extraction involved binding DNA to a white silica gel membrane, washing by transferring it to different collection tubes, and spinning them, a process repeated twice. A buffer was used to separate the DNA extract from the column.
In PCR, a reaction mix was prepared sufficiently for all the samples and surplus just in case of errors during pipetting. New tubes were labeled, and an additional tube for the positive control was provided as an extract from the species in the study. Reagents included buffer, magnesium chloride, dNTPs, two primers, double distilled water, enzyme polymerase Taq, and DNA. The samples were run in thermocyclers following a given program.
Results
Table 1. table representation showing total findings of the presence of Great Crested Newt from samples taken from different ponds in the Wicken Fen Natural Reserve.
| positive | negative | Unreliable | |
| Pond 1 | 7 | 0 | 11 |
| Pond 2 | 5 | 2 | 11 |
| Ditch 1 | 3 | 5 | 10 |
| Ditch 2 | 5 | 2 | 11 |
| Private pond | 7 | 0 | 11 |
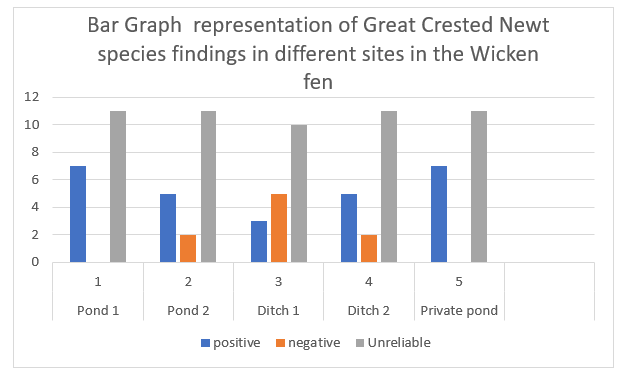
As represented in the bar graph, most of the results could have been more reliable through all the collection sites. This is shown by the tallest grey bars.
Table 2. table representation showing total findings of the presence of Great Crested Newt from samples taken during different times in the Wicken Fen Natural Reserve.
| April | may | June | ||
| Positive | 9 | 7 | 11 | |
| Negative | 4 | 3 | 2 | |
| unreliable | 17 | 20 | 17 |
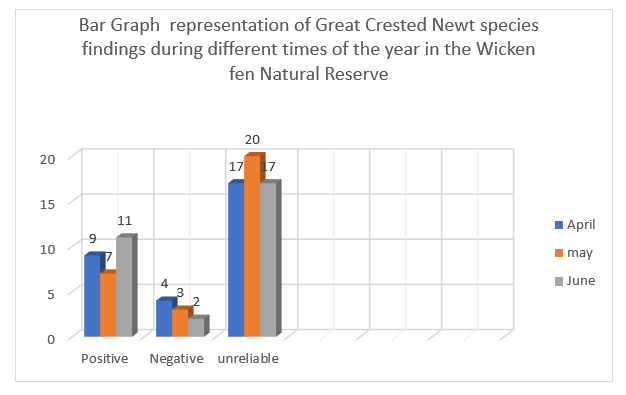
Unreliable results were the highest from all samples taken at the different months of the year at the Wicken Fen Natural Reserve, shown by the tallest bars.
Discussion
The size of the DNA fragment(s) in a particular sample and the number of distinct DNA fragments present can be determined using agarose gel electrophoresis.
DNA is present in the environment as it is shed or left over by organisms. It can be used to visualize the species’ presence in a spectrometer after electrophoresis. Still, processes including purifying and amplification using PCR must be done to concentrate the DNA.
In the practical, samples taken from the Wicken Fen natural reserve were used to determine the presence of the great crested Newt and amphibian bacteria.
The samples provided were taken systematically from two untested ponds, two untested ditches, and a pond positive for the crested Newt. The samples were taken from different pond parts to ensure sampling uniformity, then homogenized before splitting over for testing by the different groups.
The different causes of eDNA false positive and false negative findings are made clear by the results of the groups performing the same practice. Mistakes can arise either because of issues with the execution of the eDNA method or because of issues with the eDNA technique itself (method errors). Contamination of water samples with eDNA can lead to false positive detections. False negatives have been studied extensively, but some scientists have proposed solutions like using more water in the filtration process or using numerous filters.
Although this investigation did not aim to diverge from the standard Natural England procedure, future research may wish to investigate ways to optimize the gathering regimes. Similar studies of additional ponds across the UK are needed to prove how applicable these findings are to the remainder of the UK. Great crested newts may experience different breeding circumstances from year to year. Analyzing samples from multiple seasons is essential to prove this eDNA detection can be replicated.
Particularly in uncommon or threatened species, detecting a species through eDNA and field research is likely inaccurate. It may contribute to an underestimation of the spread of a species (Pilliod et al., 2013). Survey results could be skewed because many species are hard to spot at certain times of year or phases of development. Thus, the prevalence of the crested Newt within the ponds examined was likely underestimated in the data provided here. More samples may be required to achieve a greater coverage rate (especially for larger ponds). This could increase the likelihood of detecting the crested Newt by allowing the discovery of locations within the pond where the animal is most likely to be found, such as near the edges of vegetation. Future studies may find it helpful to quantify the ponds they examine to conclude the relationship between pond dimensions and detection rates, which was impossible in this one. In the future, eDNA detection with droplet digital PCR (ddPCR) could lead to higher and more reliable detection rates, as promising results have recently been obtained, but at a high cost (Nathan et al., 2014; Doi et al., 2015a, b; Simmons et al., 2015).
Conclusion
In conclusion, eDNA analysis is a technique that has been employed for the detection of a wide range of aquatic species. Based on these findings, given the limited time available for eDNA surveys of the great crested Newt, I had anticipated as much. The investigation has investigated when the species is and is not present throughout the year. The data provided in the report indicates that great crested newt DNA was detectable in water samples collected monthly from two ponds during the specified months.
References
Laramie, M.B., 2013. Distribution of Chinook Salmon (Oncorhynchus tshawytscha) in Upper-Columbia River Sub-basins from Environmental DNA Analysis. Boise State University, Boise.
Great Crested Newt (no date) The Wildlife Trusts. Available at: https://www.wildlifetrusts.org/wildlife-explorer/amphibians/great-crested-newt (Accessed: March 6, 2023).
Wilson, C. & E. Wright, 2014. Using Environmental DNA (eDNA) as a Tool in Risk-Based Decision-Making. Ontario Ministry of Natural Resources, Goulais River.
Pilliod, D. S., C. S. Goldberg, M. B. Laramie & L. P. Waits, 2013. Application of environmental DNA for inventory and monitoring of aquatic species. US Department of the Interior, US Geological Survey.
Jerde, C. L. & A. R. Mahon, 2015. Improving confidence in environmental DNA species detection. Molecular Ecology Resources 15: 461–463.
Thomsen, P. F. & E. Willerslev, 2015. Environmental DNA – an emerging tool in conservation for monitoring past and present biodiversity. Biological Conservation. doi:10.1016/j.biocon.2014.11.019.Return to ref 2015 in the article.
 write
write