Abstract
Environmental microbiomes are a new hope in hunting for novel antibacterial drugs. The Unknown Power of Soil, Aquatic, and Extreme Environment Microbiomes This essay introduces the unique antibacterial properties contained in soil microorganisms. Some key mechanisms underlying antibacterial activity, including secondary metabolites and enzymes, are described. The essay further explores the applications of environmental microbiomes in drug development, raising issues and pointing to prospects. An in-depth examination of the relevant literature reveals that these environmental microbiomes may harbor a rich goldmine of antibacterial compounds.
Keywords: Antibacterial drugs, Environmental microbiomes, Secondary metabolites, Drug development, Microbial diversity
Introduction
Antibiotic resistance is a critical and growing threat to global public health, so we must find new ways of discovering novel antibacterial agents. As resistance mechanisms evolve, the traditional methods of developing antibiotics are growing increasingly inadequate. Environmental microbiomes-microbial communities found in soil, water, or extreme environments cover another promising area for discovering new antibacterial drugs.
The increasing incidence of multidrug-resistant bacterial strains demonstrates the need for new therapeutic approaches. Vast and largely unexplored reservoirs of microbial diversity Environmental microbiomes are truly bioactive treasure troves, potentially providing a way to fight antibiotic-resistant pathogens by developing novel inhibitors (Mantri et al. (2022). Unlike traditional antibiotic discovery based on screening from culturable microorganisms, the environmental microbiome includes uncultured or difficult-to-culture bacteria, providing unexplored potential for searching out novel bioactive compounds with antibacterial properties.
Intricate microbial ecosystems exist in soil microbiomes, aquatic environments, and extreme habitats under numerous differing environmental conditions. These ecosystems have developed complex relationship chains, yielding secondary metabolites and enzymes with natural antibacterial properties. These compounds isolated from the environmental microbiomes could provide a new path for developing different antibacterial drugs.
Metagenomics as a Gateway, a major route to opening the genetic potential of environmental microbiomes, lies in metagenomic approaches, which involve direct genome analysis of entire communities (Wani et al., 2022). By exploring these complex ecosystems ‘collective genomes, researchers gain clues about the enormous extent and scope of microbial life on earth and new possibilities for biosynthetic pathways to yield entirely novel antibacterial compounds. Metagenomics provides a forceful weapon for narrowing the chasm between microbial diversity and drug discovery.
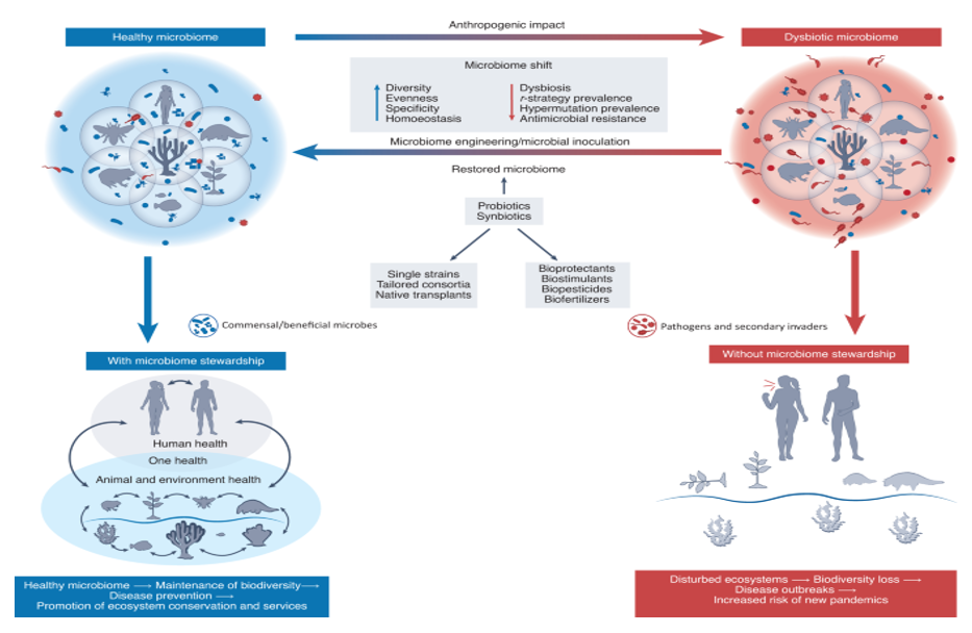
Figure 1: Microbiome stewardship as a potential tool to mitigate anthropogenic impacts
The Achilles’ heel of traditional methods for discovering new antibiotics has been the repeated rediscovery of known compounds, and most environmental microorganisms cannot be grown in laboratory conditions. Environmental microbiomes leverage their unculturable or difficult-to-culture microorganisms to address these shortcomings, opening up new frontiers in the search for highly selective antibacterials.
As the prevalence of antibiotic-resistant bacteria rises, so does the urgent need for new targeted treatments. The environmental microbiomes are an immense and unexplored treasure trove that could contain anti-resistance agents. Unique chemical diversity within these ecosystems may be the key to unlocking solutions in response to a changing landscape of antibiotic resistance.
The essay ends with one realizing the boundless possibility that environmental microbiomes represent antibacterial drug sources. It seeks to clarify exactly how these organisms work by looking at the antipathogenic properties of a soil, aquatic, and severe climate microbiome. In addition, the essay discusses many of the challenges and opportunities to make active use of these microbiomes for drug development; this requires collaboration across boundaries on various levels and technological improvement.
As individual passengers on uncharted trips to the kingdom of environmental microbiomes, we hope this trip will offer new antibiotics to fight disease and open a freshly defined route for drugs against multi-antibiotic resistance.
Antibacterial Properties of Environmental Microbiomes
Soil Microbiomes
The soil ecosystem is extremely diverse microbes and encompasses many intersecting relationships. Soil microbiomes are rich in antibacterial properties; hence, they have become a focus of research and are packed with bioactive components to be used as drug ingredients.
Richness of Secondary Metabolites
Diverse in structure and function, secondary metabolites are prolific producers of soil microbiomes. They are strong antibacterials for microorganisms in the competitive soil environment. This chemical richness of these compounds reveals the adaptability and resourcefulness of soil microorganisms. These various wonderfully shaped structures are themselves byproducts of evolutionary pressures. Structural diversity, from simple to complex molecules, enables the soil microbiome’s ecological balance (Wydro, 2022). Other than self-protection, the antibacterial properties of secondary metabolites have potential applications in agriculture and medicine as well. These compounds have potential applications in developing new antibiotics and biopesticides, tackling the problem of microbial resistance as well as many infectious diseases. They also provide stimulation for innovation throughout a wide range of scientific fields.
Antimicrobial Peptides
Dynamic defense systems Antimicrobial peptides (AMPs) occur throughout the warp and weft of soil microbiomes. These tiny yet potent peptides have antimicrobial effects that work in several ways. AMPs destroy bacterial membranes, disrupt critical cell functions, and even attack specific parts of a cell. These kinds of collective efforts surround them with our bacterial enemies. AMPs may make the soil microbiome abundant. The peptides have different sequences and work in a variety of ways. This diversity demonstrates the rough and tumble of their adaptations in this ceaseless struggle for survival, but it also offers a virtual goldmine to be mined by the biotechnologist. But can AMPs in soil microbiomes lead the way to new antimicrobial agents? Such studies all serve to point humanity’s efforts in the right direction and, therefore, are of great use for mankind’s fight against infectious diseases, as well as preventing an imminent antibiotic resistance crisis.
Enzymatic Antibacterial Activity
Antibacterial enzymes are one strong weapon used daily in the constant war of bacteria against each other. These enzymes are different types and kill bacteria by disrupting their life functions. These enzymatic agents directly target the bacterium’s life cycle stages, such as protein synthesis or cell-wall formation, which are essential to their survival. The enzymatic capabilities of the microbiome in soil comprise a diverse, high-efficiency arsenal against other microscopic organisms. Diversity reflects the adaptability of microorganisms in the ever-changing soil environment where survival depends on using a variety of strategies against other populations of bacteria and fungi vying for autophagy (Schurr et al., 2023).
Metagenomic Approaches for Discovery
Through metagenomic technologies, scientists have performed a complete health assessment on collective genetic material and revolutionized how new antibacterial compounds are discovered within soil microbiomes. This direct survey of environmental DNA allows researchers to access the genetic resources traditionally obscured by cultivation-based techniques. This method has effectively found new biosynthetic gene clusters that produce antibacterial compounds. Besides enriching our understanding of the microbial defense system, metagenomics also hastens bioactive molecule discovery. Unveiling elaborate biochemical arsenals’ metagenomic exploration, with its ability to crack the genetic code of formerly unculturable microorganisms of antibacterial agents, is another major development (Tracanna, 2022). It remains possible that the transformative method will change medicine, agriculture, and biotechnology by developing new therapeutic strategies and sustainable solutions.
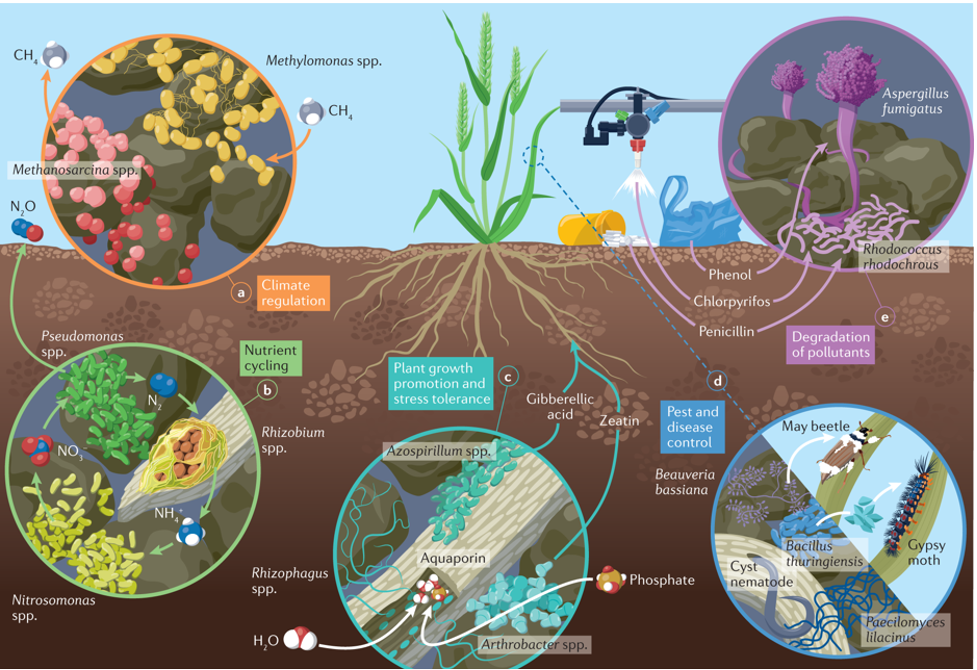
Figure 1: Antibacterial activity of soil microbiomes
Soil microbiomes are a rich resource of antibacterial compounds that produce various secondary metabolites, AMPs, and numerous enzymatic activities. These studies into these microbial communities reveal an intricate dance of chemical warfare between microbes within a single ecological niche. The metagenomic approach, combined with an awareness of the different ecological niches within soils rich in microorganisms called actinomycetes, opens up huge potential to discover new antibacterial agents.
Aquatic Microbiomes
Aquatic microbiomes include oceans, lakes, rivers, and estuaries. These harbour a wonderfully diverse array of different types of microorganisms. They are an important control point for nutrient cycling, carbon sequestration, and ecosystem balance. Recent research has claimed that aquatic microbiomes play a large part in producing bioactive compounds with strong antimicrobial activity.
Forced to fight for scarce resources in a marine environment, microorganisms have developed intricate chemical warfare techniques. Secondary metabolites, peptides, and polyketides are strategies used by many marine bacteria to get ahead in an ecological niche (Hemmerling & Piel, 2022). Such compounds are strong antibacterial agents and can be used as lead molecules in drug development.
In addition, the special environmental conditions of hydrothermal vents and cold seeps have given rise to extremophiles with exceptional adaptive capabilities in dealing with extreme pressure, temperature diffusions, or chemical gradients. These extremophiles have evolved bioactive compounds that are stable and antimicrobial simultaneously in harsh environments. Studying these extremophiles and their metabolites points to a bright path in the search for new antibacterial agents.
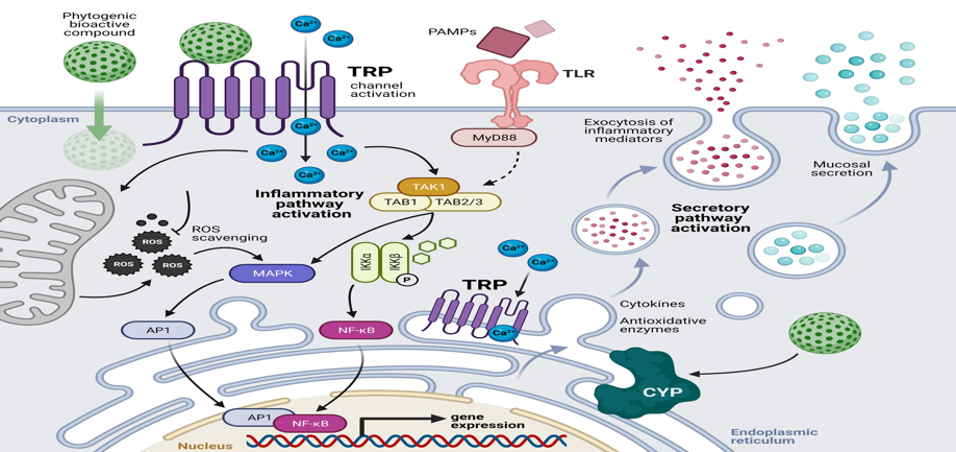
Figure 2: Diversity of bioactive compounds in aquatic microbiomes
Traditional culture-based techniques aside, metagenomic approaches have revealed a veritable genetic cornucopia among the microbiomes up and down aquatic environments. With metagenomic analyses, studying the group genomes of entire microbial communities is possible. Thus, researchers can search for genes involved in producing bioactive compounds. This all-encompassing method has greatly broadened our knowledge of genetic variation and functional possibilities in aquatic microbiomes in antibacterial drug discovery.
Yet the deeper we venture into these largely unexplored oceans of aquatic microbiomes, the greater our chances of finding new antibacterial compounds. The richness of these environments offers great potential for tackling the problem of antibiotic resistance and developing novel therapeutics.
Extreme Environments
High temperatures, acidic pH, and extreme salinity represent unique microbial life niches. But despite being thought by many organisms to be hostile places, these environments are home to the extremophiles, microorganisms adapted in their evolution for life under such extreme conditions. From extreme environments as sources of antimicrobial drugs, a mine of bioactive compounds with potential medical applications is opened.
Extreme Temperature Environments: Some are adapted to extreme temperatures, inhabiting hydrothermal vents and geysers. Unless they can withstand conditions fatal to most organisms, such extremophiles could not survive. Heat-stable enzymes and secondary metabolites produced by these extremophiles may have robust antibacterial activity (Ibrahim et al., 2023). It is of particular interest for their stability at high temperatures, which makes them useful in industrial processes and, possibly as therapeutic agents, are enzymes from extremophiles -including thermophilic bacteria.
Extremophiles that produce bioactive compounds with strange properties live in acidic environments, like acid mine drainage sites, and alkaline ones, like soda lakes. The high pH conditions serve as a selective pressure, causing the production of antimicrobial substances. Environmental compounds may destroy the cell membrane and other vital parts of bacterial cells.
Microorganisms that tolerate high salt concentrations found in salt flats and hypersaline lakes synthesize compounds rich enough to combat stress. The compounds are often antimicrobial, making them interesting candidates for developing into drugs. Potentially, halophiles could furnish compounds that damage bacterial cell walls or interfere with critical cellular activities.
These extreme environments also contain bioactive chemicals not typically found in the chemical structures of non-extreme ones. In the fight over antibiotic-resistant strains, these compounds might even affect bacteria differently than other weapons. Studies into extreme environments help researchers make a breakthrough in the chemical structure of potential antibacterial substances.
Applications of extremophiles and bioactive substances are not confined to the production of drugs. The enzymes of extremophiles, used in several industrial processes, are a perfect example. Due to their special features, they make wonderful catalysts used in biocatalysis or otherwise. Apart from this application, there are other possibilities for extreme environment microbiomes.
Figure 3: Microbial adaptations in extreme environments
Severe environments, too, require methods of sampling and cultivation. However, through metagenomic analyses of the biodiversity among extremophiles and their bioactive compounds, scientists can offer people another vision of life. Hence, these extreme environments constitute an ideal testing ground for antibacterial agents. Developing extreme environments by domesticating such microorganisms gives us a novel range of bioactive molecules with promising chemical structures. Exploring these worlds of extremity helps to better comprehend microbial life and gives channels to develop new antibiotics.
Mechanisms of Antibacterial Activity
Secondary Metabolites
Microorganisms produce secondary metabolites to mediate their antibacterial activity. These compounds, found in environmental microbiomes (particularly soil), form a family with distinct structures and mechanisms that allow them to be antibacterial.
Chemical Diversity and Structural Complexity
Soil-derived secondary metabolites Examples of splendid chemical diversity include the microbiome. These compounds are synthesized by microbial biosynthetic pathways, the structure of which is extremely complex. This variety means there are ample opportunities to attack bacterial pathogens. It has broad antibacterial activity.
Inhibition of Cell Wall Synthesis
Most secondary metabolites act by inhibiting the formation of bacterial cell walls. They inhibit the development of an important element–peptidoglycan bacterial cell walls. The microorganisms found in the environmental microbiome have an effect similar to antibiotics by breaking down the bacterial cell wall and preventing the proliferation of pathogenic bacteria.
Disruption of Membrane Integrity
Another common mechanism of the secondary metabolites is to disrupt bacteria cell membranes. They may also percolate the lipid bilayer and drain segments of intracellular content, resulting in cell death. This action is especially potent against various varieties of bacteria and complements environmental microbiomes ‘antimicrobial effects.
Inhibition of Protein Synthesis
Bacterial protein synthesis is essential to cell survival and function, so the secondary metabolites interfere with this process. They interfere with fundamental cellular processes by acting on ribosomes or some other part of protein synthesis. This highly selective process can be the basis for developing antibiotics that act against specific targets.
Interference with Nucleic Acid Synthesis
Yet some secondary metabolites disrupt nucleic acid synthesis. They are also poisons but target the genetic machinery of bacterial pathogens- in other words, their replication processes for either DNA or RNA. Such behavior is especially effective at suppressing bacterial proliferation and preventing the spread of genetic information, making environmental microbiomes a potent weapon against bacteria.
Biofilm Disruption
Biofilm-disrupting properties may reside in the secondary metabolites of environmental microbiomes. Bacterial biofilms are complicated structures that can lead to resistant bacteria and chronic infections. Strategies targeting biofilm formation or destabilizing established biofilms provide valuable ways of fighting persistent bacterial infections.
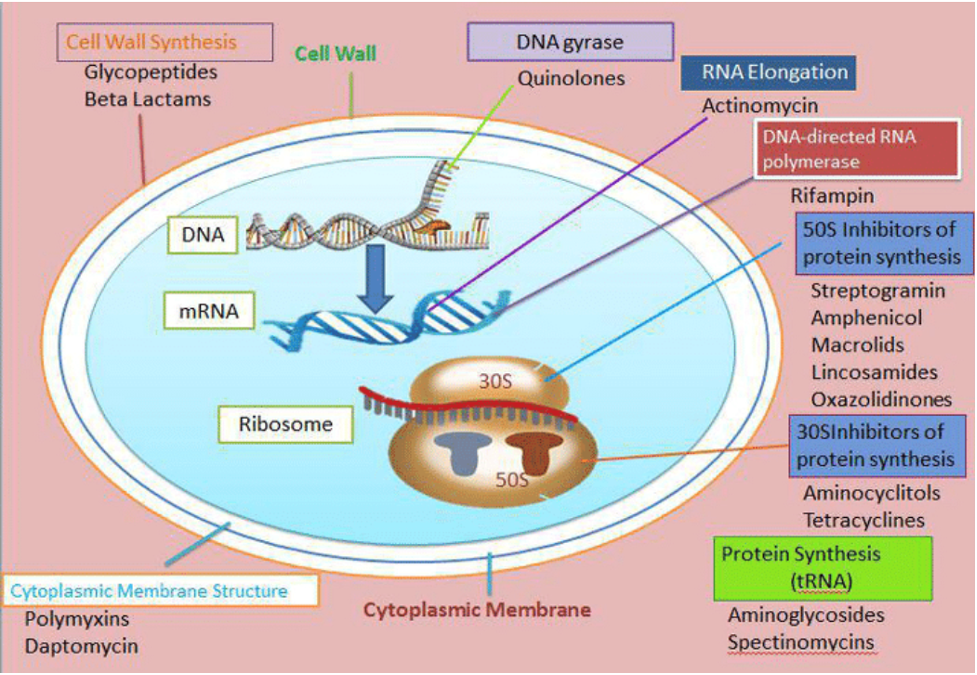
Figure 2: Modes of action of secondary metabolites in antibacterial activity
In other words, the numerous secondary metabolites produced by microorganisms within environmental microbiomes demonstrate several antibacterial mechanisms. Spanning the inhibition of cell wall synthesis and disruption of membrane integrity to interference with essential cellular processes, these compounds remain a rich reservoir for new antibacterial agents possessing widely different modes of action.
Enzymes
Enzymes secreted by the microbes in environmental microbiomes are a big part of our antibacterial weapons. Some of these biocatalysts, often with highly specific targets within bacterial cells, lend to the myriad mechanisms by which environmental microbiomes fight off pathogenic bacteria.
Cell Wall-Degrading Enzymes
Antibacterial mechanisms enzymes targeting bacterial cell walls are prominent. For instance, lysozymes can break down peptidoglycan–a fundamental component in the structure and strength of bacterial cell walls. This enzymatic activity dissolves the structural integrity of the cell wall, bacterial lysis, and abolition. The cell wall-degrading enzymes are active against many different bacterial species.
Proteases and Protein-Degrading Enzymes
Environmental microbiomes produce proteases that degrade essential proteins for bacterial survival, thus leading to the disruption of bacterial homeostasis. These enzymes may act on structural proteins, cellular process-related enzymes, or virulence factors. The result is that bacterial growth will be inhibited and pathogenicity attenuated.
Nucleases
Other enzymes, such as DNases and RNAases, contribute to the antibacterial activity by destroying nucleic acids inside bacterial cells. These enzymes could compromise the integrity of DNA or RNA, impairing important genetic functions and destroying bacterial reproduction. Nucleases are especially potent in reducing the risk of genetic material spreading and limiting bacterial contagions.
Lipases and Lipid-Degrading Enzymes
By hydrolyzing lipids, Lipases and other lipid-degrading enzymes attack the membranes of bacteria. Lipid-bilayer disruption compromises membrane integrity and leads to leakage of intracellular components. The bacteria die as a result of this breach in their cell walls. Environmental microbiomes rely on lipid-degrading enzymes to enhance their overall antibacterial ability.
Quorum Sensing Inhibitors
Some enzymes inhibit bacterial communication systems sensing. Quorum sensing is critical for coordinating bacterial activities, such as biofilm formation and expression of virulence factors. Quorum-sensing pathways are disrupted by enzymes, which can prevent the establishment of biofilms and reduce bacterial virulence.
Oxidative Enzymes
Oxidative enzymes such as catalases and peroxidases produce reactive oxygen species (ROS) inside bacteria. The ROS oxidizes cellular components, killing the bacterial cells. Environmental microbiomes fight off bacterial pathogens by seizing the opportunity when bacteria experience oxidative stress.
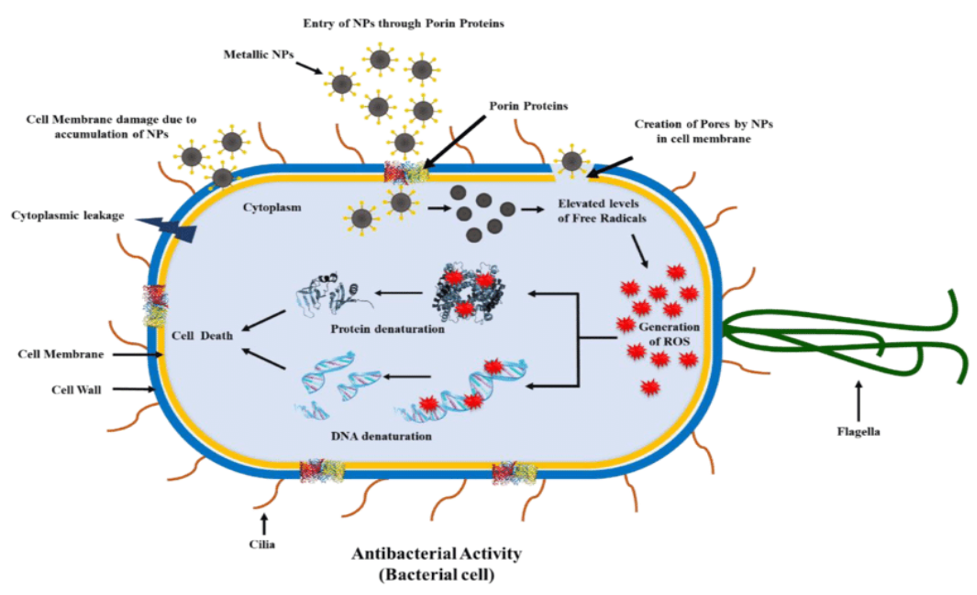
Figure 3: Enzymatic mechanisms in antibacterial activity
In sum, the enzymatic weapons within environmental microbiomes (soil as an example) are a thousand different biocatalysts aimed at specific parts of the bacterial cells. They provide complex and highly specific mechanisms for destroying bacterial pathogens. Every conceivable keyhole has been fitted in preparation, from enzymes that dissolve cell walls to those that cleave proteins, interfere with nucleic acids, or rupture membranes.
Applications in Drug Development
The environmental microbiomes can have applications in almost every stage of the pharmaceutical pipeline. In particular, they provide many starting points for developing new antibiotics and reducing resistance to current drugs.
The environmental microbiomes provide many sources for identifying good antibacterial lead compounds. The rich array of chemical structures among the secondary metabolites and enzymes that microorganisms produce within these ecosystems represents a veritable mine of leads. Systematic screening and characterization allow researchers to identify compounds acting in novel ways. From there, the active compound can be further developed.
When lead compounds are established, hit-to-lead optimization is an important stage in drug development. This means purifying and transforming the chemical characteristics of effective compounds to strengthen their pharmacological features, such as activity, specificity, and absorption. The properties of chemical scaffolds are diverse in the environmental microbiome, providing rich ground for optimization.
The concept of environmental microbiomes may help the development of combination therapies, which are designed to use several drugs with different mechanisms to better combat bacterial infections (Sorbara & Pamer, 2022). If researchers can harness all the diversity of antimicrobial compounds within these ecosystems, they can explore synergistic effects and overcome single-drug resistance. Using this method may increase the effectiveness of treatment and prolong the life span of antibacterial drugs.
Such strains pose a public health problem. Environmental microbiomes abound with bioactive compounds, making them an excellent place to cultivate agents targeted at resistant strains. Understanding how these drugs work enables researchers to develop new therapeutics with which we can face a constantly shifting antimicrobial resistance situation.
In hospitals, biofilms are a serious threat. Make chronic infections common and make it easy to fight the enemy. At other times, environmental microbiomes (typically featuring numerous interactions between various micro consortia) contain substances antagonistic to biofilm. Examining these exchanges provides avenues for developing drugs against biofilm-associated infections.
Challenges and Future Prospects
As exploration into the microbiomes of our various environments continues to grow, biodiversity conservation and sustainable use have become increasingly relevant ethical issues. Ethical requirements are also held concerning sample collection, and investigations should not harm the ecosystems of microbiome origins.
The prospects for environmental microbiome-based antibacterial drugs are intimately bound up with advances in metagenomics, synthetic biology, and bioinformatics. Advances in these areas of innovation will enable us to continue extending our knowledge of the potential within microbial communities. To the extent that it is possible today to accurately identify strains, isolate them, and have 100 % effective production methods. If such problems can be resolved, this would revolutionize world food safety standards.
Environmental microbiomes also represent tremendous potential for hunting out new antibacterial drugs. These obstacles will only be overcome with the combined efforts of researchers, industry partners, and relevant authorities. The environmental microbiomes that harbor such rare substances may one day become a source of precious quantities of novel antibacterial compounds that can truly overcome drug resistance.
Conclusion
Using environmental microbiomes in antibacterial drug development is a new approach to addressing this antimicrobial resistance problem. There are many bioactive substances in microbiomes found in soil, aquatic, and extreme environments that can be turned into drugs. Nevertheless, by studying the principles of antibiotic action and resolving these issues, scientists can uncover enormous potential lying dormant in microbial communities. If environmental microbiomes are not introduced into the drug discovery process, we will be unable to win this war against antibiotic resistance, and our health won’t have a secure future.
References
Hemmerling, F., & Piel, J. (2022). Strategies to access biosynthetic novelty in bacterial genomes for drug discovery. Nature Reviews Drug Discovery, 21(5), 359-378.
Ibrahim, K. S., Aishwarya, M., & Kannan, R. P. B. (2023). Secondary metabolites from extremophiles with therapeutic benefits. In Recent Advances and Future Perspectives of Microbial Metabolites (pp. 249–267). Academic Press.
Mantri, S. S. (2022). Metagenome mining explores novel regions of natural products’ chemical space (Doctoral dissertation, Universität Tübingen).
Schurr, U., Pinheiro, C., Fasoula, D. A., Goulas, E., Carpentier, S. C., & Junker, A. (Eds.). (2023). Phenotyping at Plant and Cell Levels: The Quest for Tolerant Crop Development, volume II. Frontiers Media SA.
Sorbara, M. T., & Pamer, E. G. (2022). Microbiome-based therapeutics. Nature Reviews Microbiology, 20(6), 365-380.
Tracanna, V. (2022). Dissecting disease-suppressive rhizosphere microbiomes using metagenomics (Doctoral dissertation, Wageningen University and Research).
Wani, A. K., Roy, P., & Kumar, V. (2022). Metagenomics and artificial intelligence in the context of human health. Infection, Genetics, and Evolution, 100, 105267.
Wydro, U. (2022). Soil microbiome study based on DNA extraction: A review. Water, 14(24), 3999.
 write
write