Introduction.
Induced Pluripotent Stem Cells (iPSC) are cells derived from the skin or blood cells that have been reprogramed back into the embryonic-like pluripotent state that facilitates the development of an unlimited source that can be repurposed into any other type of cells needed for other purposes, such a treatment. iPSC is typically derived by introducing a specialized set of pluripotency-associated genes or reprogramming factors into an adult somatic cell to induce differential (Seaby et al., 2015). The iPSC cells have qualities similar to human embryonic stem cells. According to (Li et al., 2015), the initial set of reprograming factors, also called the Yamanaka factors after Professor Shinya Yamanaka, established their potency in 2006. The Yamanaka factors are (Oct3/4, Sox2, Klf4, and c-Myc) (Li et al., 2015). They are described as a group of protein transcription factors responsible for forming pluripotent stem cells, capable of transforming into any other cell in the body. The cells control how DNA is transferred and eventually transformed into other protein cells. The discovery of the Yamanaka factors was a crucial development in stem cell research and offers a possibility for understanding cell transformation factors that allow researchers to repair broken cells or create new cells, thus making limitless possibilities in treating diseases and other conditions.
Heterogeneity of Induced Pluripotent Stem Cells (iPSC).
Heterogeneity refers to the existence of different characteristics in a group. In cell biology, heterogeneity expresses other traits from cells often classified in the same group. When referring to Induced Pluripotent Stem Cells (iPSC), line/clones’ heterogeneity refers to the genetic difference between the single iPSC clones that manifest after mutation of the somatic cells that arise after the cell is subjected to various treatments (Seaby et al., 2015). According to (Li et al., 2015), numerous mutations may result in the heteronomy of the iPSC clones. The different mutations manifest for a variety of reasons and can be a crucial aspect in helping researchers devise ways of determining the heterogeneity of the stromatic cells (Li et al., 2015).
The approach can be an effective avenue for determining the pluripotency of the stem cells that are included in the panel for detecting stem cells before differentiation. The common pluripotency markers that are included in a panel for detecting stem cells prior to differentiation include OCT4, NANOG, and SOX2. The study allows researchers to map the characteristics of iPSC before they differentiate (CD Genomics, 2023). Understanding the features of the stem cells before differentiation allows the researchers to map out the protein characteristics of all the elements within the cells (Seaby et al., 2015). The researchers can, therefore, utilize several markers to identify the differentiated iPSC and the differentiated iPSC (Aboul-Soud et al., 2021). The standard surface markers selected from the experience are TRA-1–60, Stage Specific Embryonic Antigen 4 (SSEA4), and TRA-1–81. SSEA-4 expression seems to precede the expression of TRA-1–60 and TRA-1–80, which are only noticeable in the final phases of differentiation. However, TRA-1–60 and TRA-1–80 are preferred because they have unique epitopes of the glycoprotein Podocalyxin and are used to identify isolated ESCs (Li et al., 2015). The suggested approach will prioritize the expression of different characteristics of normal iPSC cells and differentiated versions, which are likely to have extra protein characters, especially at the cell’s nucleotide sites. Should the proteins of the differentiated and undifferent cells have other characteristics when sequences, then there is a possibility to highlight the heterogeneity of iPSC clones (Aboul-Soud et al., 2021).
Exome Sequencing to Define the Genetic Heterogeneity of IPSC Clones.
Exome sequencing is an approach to genomic studies that sequences all the protein-coding regions of the genes in a genome (Aboul-Soud et al., 2021). The exome is essential to studying diseases because it contains all the disease-related genomic variants. Exome sequencing can be an essential avenue for determining the heterogeneity of the iPSC lines because it targets the proteinoid nature of cells (Aboul-Soud et al., 2021). The portentous nature of the different components of the cells can help in exome sequencing, thus allowing researchers to determine the heterogeneity of iPSC cells (CD Genomics, 2023). The experimental method is modeled after established genome sequencing methods that usually occur through exon enrichment. Afterwards, the resultant captured library is exposed to high-throughput, followed by massive parallel sequencing to produce millions of readings (Seaby et al., 2015). The experiment will contain a control sequence that allows the comparison of the different stem cells to showcase instances of heterogeneity. The experimental method will utilize the primary steps associated with exome sequencing.
Exome sequencing is a practical approach that has been utilized in other studies and can translate well into other experiments. The proposed research experiment is inspired by the work of (Dong et al., 2022) which acknowledges the tremendous potential tool for disease modeling, drug analysis, and other applications of iPCS.
The generation of the iPCS contains the genetic history of the cell that appeals to the reprogramming process. Even the clones derived from the same individual cells are all expected to exhibit heterogeneity (Dong et al., 2022). In the experiment, the researchers performed exome sequencing on 24 mouse iPSC clones derived from the skin fibroblasts extracted from two sites in the same mouse (Dong et al., 2022). According to the researcher, there was no difference in the coding regions of the two parental fibroblast pools, and the research showed that the clone had a unique genetic marker with a wide range of heterogeneity observed among the individual clones (Dong et al., 2022). A total of 383 iPSC variants were validated for the 24 clones (mean 16.0/clone, range 0-45). Exome sequencing works as it facilitates hybridization within the somatic cells (Dong et al., 2022). According to mRNAs hybridize with primers comprising oligos (dT) and create poly C by introducing cytosine nucleotide (Aboul-Soud et al., 2021). Afterward, the primers encompassing oligos (dG) hybridize with poly C to produce the cDNAs. The full-length cDNAs are augmented by polymerase chain reaction (PCR), leading to the manufacture of nanogram-scale DNAs for exome sequencing (Li et al., 2015).
The following 8 Steps are crucial in showing how the experiment unfolds:
- The genetic materials are collected and presented in the culture in the first step. The genetic material must contain peripheral lymphocytes.
- gDNA is then extracted from the cells using specialized extraction kits or a slating-out approach; afterward the quality of the cell is assessed.
- The DNS molecule is then fragmented by enzymatic methods, which may vary between library preparations. The enzymatic method is crucial because the experimental approach targets the nucleotide’s portentous nature to showcase the iPSC clones’ heterogeneity.
- After the fragmentation ends are repaired by eliminating the overhanging nucleotides, the ends are attached to the adaptors. The process is essential as it will likely expose the protein elements of the different portions of nucleotide. The resulting rectangular regions are important as they showcase the presence of exons in the DNA sample.
- Exome isolation is an essential undertaking in the process. Through target enrichment strategies, researchers can effectively isolate the target regions, allowing them to examine heterogeneity regions in the iPSC stem cells. The enrichment process uses physiochemical properties to differentiate between the areas of interest and the obsolete areas.
- The sample is then exposed to aqueous-phase hybridization, which captures and showcases the enriched exotic sequence by ligating fragments to biomethylated probes, which is crucial in enrichment platforms. The hybridized components are recovered by biotin-streptavidin-based magnetic bead pulldown. The irrelevant region is discarded while the exome fragments are washed in a solvent to amplify their characteristics (CD Genomics, 2023).
- The resulting exome library is sequenced using parallel technologies, producing millions of sequenced patterns.
- The raw data is aligned to the human genome reference sequence and compared. The comparison phase is crucial as it allows researchers to compare the exon of the iPSC cells to regular cells, allowing them to compare the protein characteristics of the different cells.
- Data analysis.
Photographic Summary of the Process.
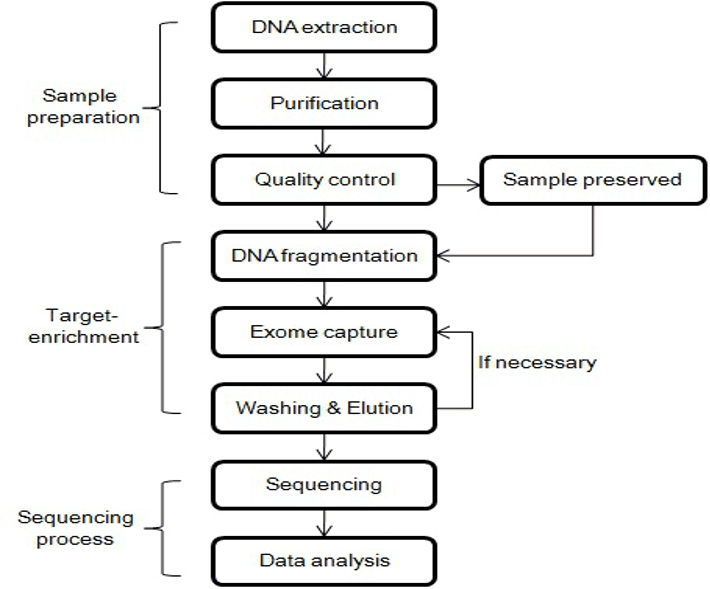
(Photo adapted from: https://www.cd-genomics.com/resourse-principles-and-workflow-of-whole-exome-sequencing.html#:~:text=Principles%20and%20Workflow%20of%20Whole%20Exome%20Sequencing%201,%20Sequencing%20technology%20…%205%20%20Data%20analysis).
Limitations and Disadvantages of the Experimental Approach.
The approach assumes that there will be numerous differences between the protein behaviors of the nucleotide cells between the differentiated and undifferentiated cells. An exome sequencing approach that highlights the protein changes is limited to the scope of characterizations of the heterogeneity it can express. The approach also does not acknowledge that cells can undergo differentiation and genetic mutation that significantly alter their behavior without changing the protein elements.
iPSCS requires a prolonged time to reach the final product. The experiment is time-consuming and utilizes numerous reagents and reactions that take time before they produce the final product. The time needed to execute the experiment can create concerns among researchers who are used to fast automated approaches.
The process is machine-dependent as several aspects of the experiment require automation. The need for machines makes it difficult to experiment with a simple laboratory experiment.
There are also concerns that the cells have a predisposition to developing cancer that can alter the effectiveness of the research. According to (Li et al., 2015) previous work from our lab suggested that many of the mutations within iPSCs actually pre-exist in rare cells within the starting cell population. The realization shows that even though the investigation can highlight the heterogeneity of iPSC, it does not reflect the possibility of the roles of mutation in the development feature (Seaby et al., 2015).
The experiment requires an expensive, controlled environment. Securing the reagents and establishing the proper lab environment for the experiment is crucial. The cost of ensuring the set-up is up to standards and the necessary observation is facilitated. With the proper infrastructure to facilitate the research, it is likely to yield good results.
There is a risk of mutation and other external factors affecting the differentiation process, which can increase the experiment errors when mapping the heterogeneity of the iPSC cells. It is crucial to find ways of reducing the venues where the errors are likely to emerge before they are addressed. However, generation is still a relatively unknown subject, and there might be light interventions the researchers can utilize to increase accuracy.
Alternative Methods.
There are other ways of using exome sequencing to determine heterogeneity in cells. Other potent approaches include single-cell epigenetic sequencing. Epigenetics refers to the alteration in gene expression levels resulting from non-gene sequence changes that showcase the functional genomic elements that can highlight the cell’s behavioral history.
Another viable approach is using Tag-based sequencing. The approach uses UMIS to characterize multiple samples with improved throughput, which is helpful for gene expression. However, the approach is insensitive because the mappable reading is restricted to one end of the cell loci.
Conclusion.
In conclusion, there are many benefits to derived from exome sequencing. Exome sequencing allows researchers to understand the gnome and its development and map how it operates and its reactions under specific treatments. Heterogeneity in the somatic cells can help scientists understand how they contribute to developing pathogens and how to respond effectively to the changes. Determining the heterogeneity of iPSC researchers can contribute to cancer research, thus saving lives. It is important to note that exome sequencing is an effective method to help determine the heterogeneity of iPSC clones/lines. However, numerous ways, both established and development approaches like array-based prosed, can also cater to the determination of heterogeneity of iPSC lines/clones. With technological advancements, scientists continue to make strides to expand the field and expose humanity to new possibilities.
References.
Li, C., Klco, J. M., Helton, N. M., George, D. R., Mudd, J. L., Miller, C. A., Lu, C., Fulton, R., O’Laughlin, M., Fronick, C., Wilson, R. K., & Ley, T. J. (2015). Genetic heterogeneity of induced pluripotent stem cells: results from 24 clones derived from a single C57BL/6 mouse. PloS one, 10(3), e0120585. https://doi.org/10.1371/journal.pone.0120585
Dong, Z., Wang, Y., Yin, D., Hang, X., Pu, L., Zhang, J., … & Chang, L. (2022). Advanced techniques for gene heterogeneity research: Single‐cell sequencing and on‐chip gene analysis systems. View, 3(1), 20210011. https://onlinelibrary.wiley.com/doi/10.1002/VIW.20210011
CD Genomics. (2023). Principles and Workflow of Whole Exome Sequencing – CD Genomics. Www.cd-Genomics.com; CD Genomics: The Genomics Service Company. https://www.cd-genomics.com/resourse-principles-and-workflow-of-whole-exome-sequencing.html#:~:text=Principles%20and%20Workflow%20of%20Whole%20Exome%20Sequencing%201
Aboul-Soud, M. A. M., Alzahrani, A. J., & Mahmoud, A. (2021). Induced Pluripotent Stem Cells (iPSCs)-Roles in Regenerative Therapies, Disease Modelling and Drug Screening. Cells, 10(9), 2319. https://doi.org/10.3390/cells10092319
Seaby, E. G., Pengelly, R. J., & Ennis, S. (2015). Exome sequencing explained: a practical guide to its clinical application. Briefings in Functional Genomics, 15(5), 374–384. https://doi.org/10.1093/bfgp/elv054
 write
write