Summary
This report entails the phylogenetic assessment of the cytochrome b gene among apes and baboons. The report is about the phylogenic assessment of cytochrome B gene among seven apes and baboon which is an Old-World-Monkey. The seven apes are; human, common chimpanzee, pygmy chimpanzee, gorilla, Bornean orangutan, Sumatran orangutan, and Gibbon.
Introduction
The most extensively sequenced gene is cytochrome B which plays a key role in the structure within the cells that are the mitochondrion. It’s a complex III protein that is involved in oxidative phosphorylation where a compound adenosine triphosphate is synthesized from oxygen and simple sugars (Hameed, 2020). Among the apes and the baboon the study of their phylogenetic cytochrome b to understand the evolutionary relationships and development over time.
Data and methods
Sequences and Alignments
The complete cytochrome B sequences were obtained from the gene bank available in their database. The sequences from this gene were obtained from the mitochondrial genomes which are complete (Garamszegi, 2016). All the sequences from accession numbers X93334,X93335, D38116, D38116, X93347, D38115, X97707, X99256, Y18001.In each of the species, one sequence was selected and one alignment was generated. During phylogenetic analysis, a common part was identified and used because these sequences are of different lengths and therefore can be hard to perform a phylogenetic analysis (Smith, 2013). The binomial names of the species were then used to name and label the sequences as it appears in the databases. Sequence alignment was aligned using CrustalW which is present and available in the MEGA software package. This approach involves a process where sequences are aligned by it being attached to a tree essentially a binary one (Garamszegi, 2016). This tool adopts a progressive approach where possible alignments were calculated and recorded, a tree was also calculated, related sequences were identified, the sequences were then aligned progressively, gapped sequences were used to extend sequences that are in agreement, and lastly the multiply aligned sequences were reported. A potential problem to this alignment process is the increase in the sequences numbers which is a current development.
Phylogenetic Analysis and Trees
The phylogenetic tree contains nodes where ancestral or descendant lineages are generated.it has various branching points which represent the transfer of genetic data from one generation to another (Smith, 2013). The parts of a tree are symbolic in a way where the common ancestor is represented by the root, the leaves represent species, genes, and individuals and the branches are the regions for passing genetic information.
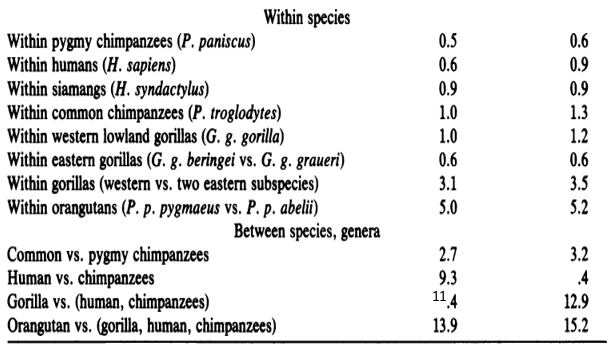
Table 1. below shows sequence differences between the primates
Cyt b sequences of the species were developed and constructed using Maximum likelihood methods and MrBayes. A calculation was done using MrBayes and RAxML then the data obtained was exported online and given different colors to enable for good visual presentation (Wünschiers, 2013). RAxML a method that was used to calculate the maximum likelihood approach uses rapid bootstrapping. Bootstrapping is used in assessing the accuracy of most statistical values.
This method has been applied in the estimation of phylogenetic trees. The code works through a sampling of random bases having a replacement and new sequences are made (Smith, 2013).To make a pairwise alignment, the gaps are removed with the same number of bases used. A function by the name and sample samples data has a replacement although the function can also sample the data randomly without replacement to perform analysis. For this type of analysis called jackknife, there is a replication which forms a repetition of each sequence. With up to 100 repeats the bootstrapping was performed afterward (smith, 2013).100 repetitions is a reversible design of a nucleotide substitution having four rate categories. The trees to be calculated using the MrBayes approach were set to 4 by 4 nucleotide model which is a type of DNA type. The other things set were Dirichlet frequencies of the four states and an invariable gamma which is within the default settings.
| 156 | 177 | 204 | 267 | 268 | 342 | 414 | 525 | Sd9 | 6l3fi | 637 | ||
| Hsa1 Hsa2 Hsa3 Hsa4 | T | T | G | G | G | G | C | T | T | T | A | T |
| Hsa6 | ||||||||||||
|
Pw3 Pn4 |
||||||||||||
|
Ppa4 |
||||||||||||
| Ggo1 | A | C | A | A | A | A | T | C | C | A | C | C |
| Ggo2 | A | C | A | . | A | A | T | C | C | A | C | C |
| Ggo3 | A | C | A | A | A | A | T | C | C | A | C | C |
| Ggo4 | A | C | A | A | A | T | C | C | A | C | C | |
| Ggo5 | A | C | A | A | @ | A | T | C | C | A | C | C |
| Ggo6 | A | C | A | A | A | A | T | C | C | A | C | C |
| Ppy1 | G | C | A | A | A | A | C | C | A | T | C | |
| Ppy2 | G | C | A | A | A | A | C | C | A | T | C | |
| Ppy3 | G | C | A | A | A | T | C | C | A | C | ||
| Hsy1 | A | C | A | A | A | A | T | C | C | A | C | C |
| Hsy2 | A | C | A | A | A | A | T | C | C | A | C | C |
Table 2. Shows the gene variations among humans and the other apes is an example of the relationship of evolutionary development of the species.
Results and discussion
The results of the Bayesian software which was able to estimate the divergence and the phylogenic tree show that we agree with the studies which show relationships across all the primates that is apes and baboons (Hameed, 2020). The construction of phylogenetic trees of the separate trees and the subsequent sequence analysis demonstrated the intragenic and intergeneric distances between the species. There is a variation which is systematic of the cytochrome b on factors such as body size this is due to the differences in evolutionary rates of the gene within the primates under study (Hussain, 2020). As the lineages evolve among primates there is a distinct difference within the gene. Study shows that there are reasons for example generation time and metabolic rate influences their respective body mass and is assumed to be responsible for the difference.
The figure above shows that there may be a relationship between cytochrome b divergence and the body mass of these organisms. Among small animals, evolution rates of cytochrome b are increased. Through comparisons of phylogenetic pairs of primates on the trees, it shows that there is no basis between the maximum likelihood ratios and body mass. This illustrates that the relationship between body mass and cytochrome b cannot explain the increase in the evolution rate among small primates (Hussain,2020). This study is regarding both taxonomic and gene study of the species which are primates. Genomic sequences illustrate a similar phylogenetic tree because of the existence of overlapping parts of the amplified gene.
From other molecular studies, divergences among baboons are very consistent. Baboons have changed in size due to partial distribution and periodical climatic changes in their habitats. Among the Homo sapiens, gene flow is confirmed to have taken place over time among populations illustrating the cyt b phylogenic differences. Aspects for example glacial changes for different migration routes in the case of African content.
Conclusion
In conclusion, cytochrome b has greater correlations with the phylogeny of the primates together with having greater variations in shorter sequences of the gene. Despite the differences in body sizes of these species, there is a generic distance within the genomic sequences of the cytochrome b gene. This assessment of genes can be used to study the evolution stages and development among different species. Through this report, we can identify the differences among species. The Old-World-Monkey, for example, has undergone several changes within the cytochrome b gene over the years and we can use it to study its evolutionary development.
REFERENCES
Khalid, M.; Hussain, T.; Farooq, Z.; Abbas, K.; Babar, M. E. Mitochondrial Cyt-b and Cox1 Genes Based Molecular Diversity and Phylogenetic Analysis of Chukar Partridge (Alectoris Chukar). Pakistan Journal of Zoology 2020, 52 (4).
Baum, D. A.; Smith, S. D. Tree Thinking: An Introduction to Phylogenetic Biology; W.H. Freeman And Company: New York, NY, 2013.
Röbbe Wünschiers. Computational Biology: A Practical Introduction to Biodata Processing and Analysis with Linux, MySQL, and R; Springer: Heidelberg; New York, 2013.
Hameed, U. Importance of Cytochrome Oxidase Subunit 1 in Phylogeny Reconstruction of Various Classes of Subphylum Vertebrata Usman Hameed. Pure and Applied Biology 2020, 9 (3).
László Zsolt Garamszegi; Springer-Verlag Gmbh. Modern Phylogenetic Comparative Methods and Their Application in Evolutionary Biology Concepts and Practice; Berlin Springer, 2016.
 write
write